We have recently developed a novel informatic toolbox applicable to any species or geographic area to predict vulnerability to global change, ultimately aimed at improving conservation prioritisation and monitoring efforts. You can find the published manuscript here and the DRYAD and ZENODO repositories for data and code here. Whenever I update code it will be available through the LOtE github repository and linked in the LOtE news. If you encounter any issues with the code, or want to collaborate please contact me (c.d.barratt@gmail.com), but do please note that I do have a full time research position with many other responsibilities, so it may take time for me to get back to you.
The toolbox facilitates the integration of environmental (e.g. climate, land use), ecological (e.g. spatial occurrences), and evolutionary (e.g. genome-wide SNP) data via a series of modular scripts and functions. The toolbox can be run from start to finish (i.e. raw spatial, environmental and genomic data) through to final population vulnerability maps), or specific modules can be used separately (e.g. if you just want to build Species Distribution Models, look at population striucture or perform Genotype Environment Association analyses).
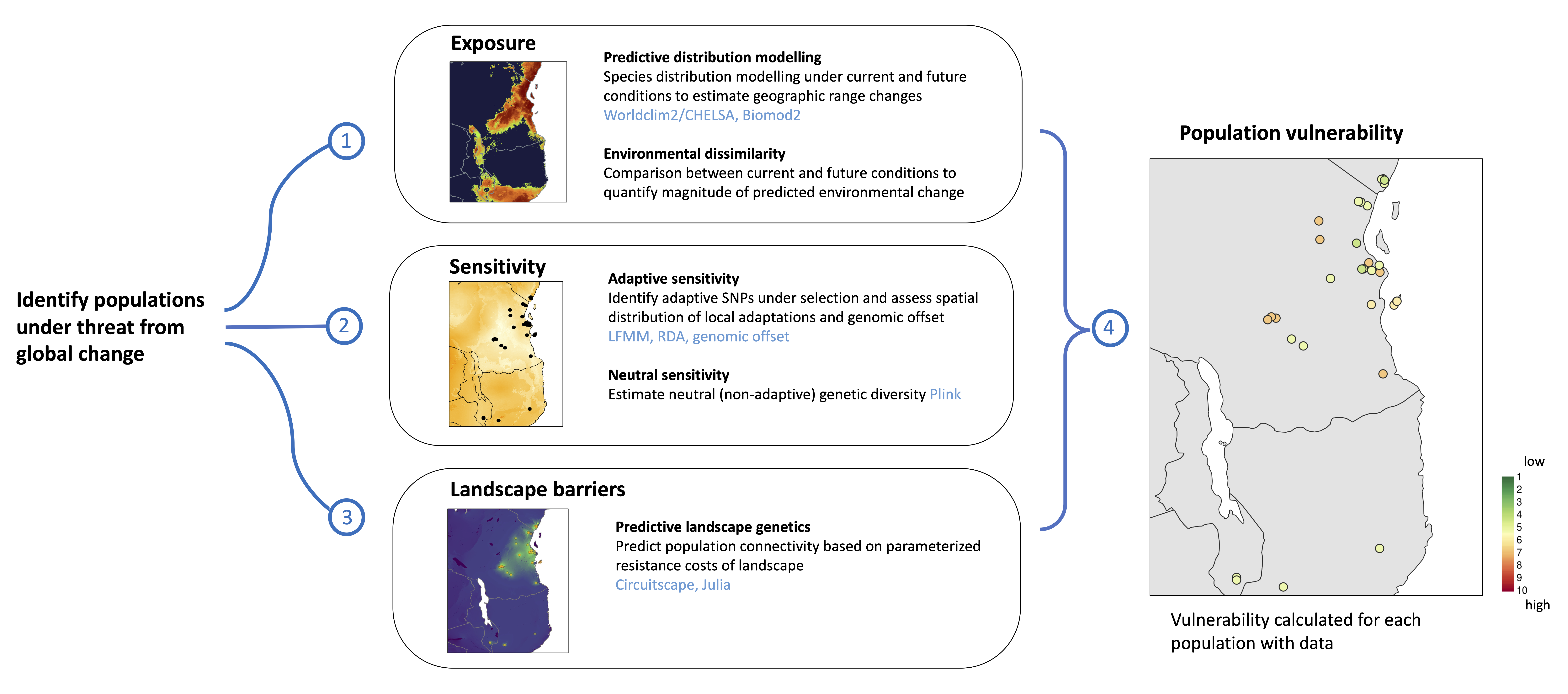
We follow the frameworks of two main papers, Razgour et al. 2018 and Razgour et al. 2019, via a series of scripts and functions that have been generalised with flexible code to accomodate any species input data from any geographic region. The toolbox estimates four main metrics for each population/sampling locality:
- Exposure - the magnitude of predicted change at future climatic conditions (i.e. environmental dissimilarity + changes in habitat suitability using species distribution models)
- Neutral sensitivity - the neutral sensitivity of each individual/population to global change based on genomic data (i.e. lower nucleotide diversity = higher neutral sensitivity)
- Adaptive sensitivity - the adaptive sensitivity of each individual/population to global change based on genomic and environmental data (i.e. high genomic offset = higher adaptive sensitivity)
- Landscape barriers - the landscape connectivity resistance for populations to be able to move (i.e. based on current landscape connectivity)
These four metrics are then assessed to calculate final Population vulnerability (see figures below for workflow and final output)
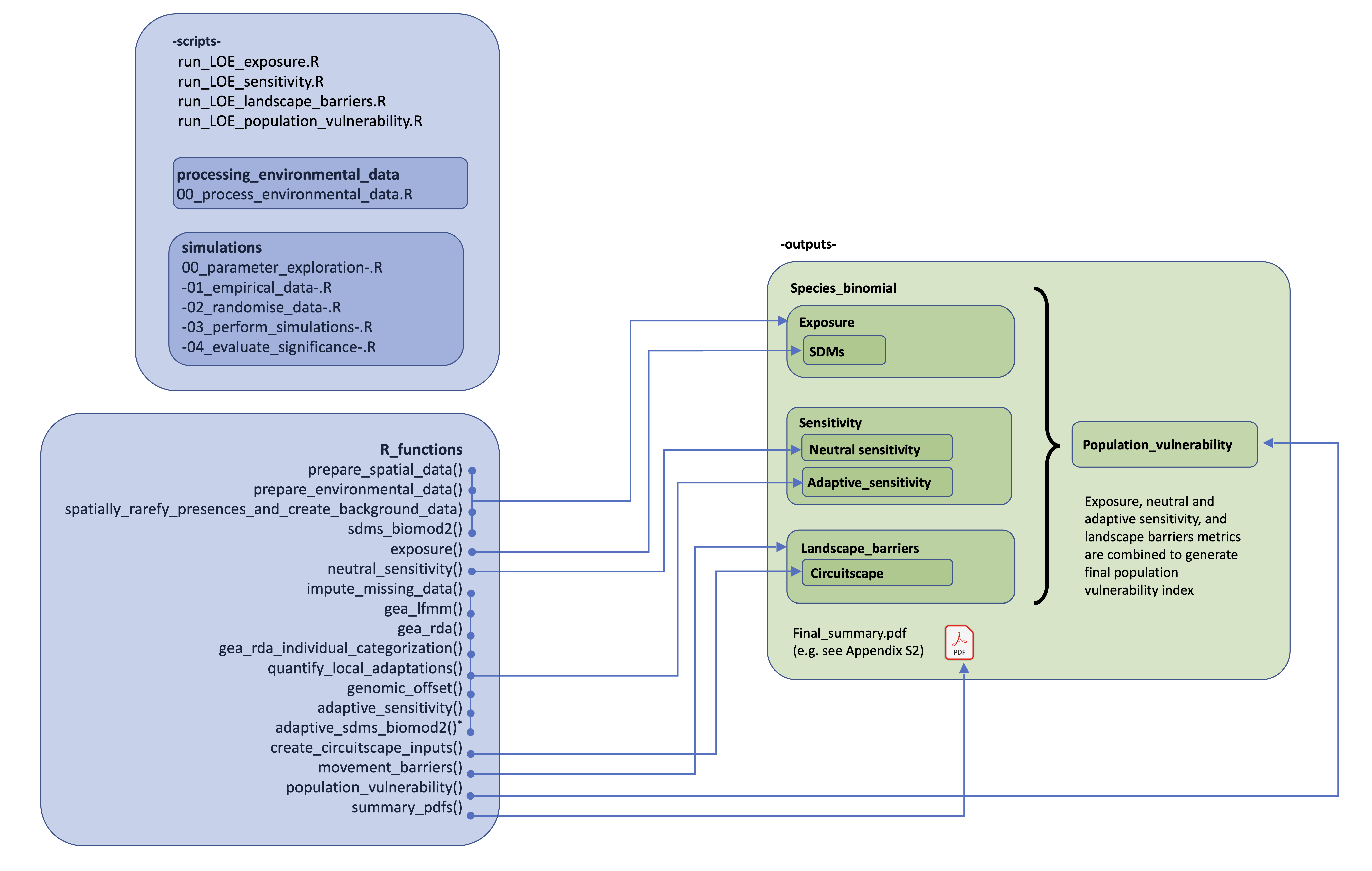
The toolbox runs from a Params.tsv file (up to 71 parameters which may be defined/modified), and all you need to provide are the spatial, enviromental and genomic input data per species (or specify do auto-download the enviromental data from Worldclim)
Below a list of some of the things you can do using the Life on the edge toolbox:
- Download and process spatial data (e.g. GBIF and georeferenced genomic data), clean it and prepare environmental data (e.g. Worldclim) for Species Distribution Modelling (including clipping to relevant extents per species, spatially rarefying input data to control for spatial autocorrelation, and preparing background pseudoabsence data)
- Run ensemble species distribution models and predict future changes based on user specified parameters
- Calculate the magnitude of environmental change predicted to occur for each population (based on SDMs)
- Perform Genotype-Environment Association Analyses with user specified environmental predictors (RDA, LFMM)
- Use simulations and sensitivity analyses to statistically validate the candidate SNPs you have identified using permutation tests
- Individually categorise individuals in GEA ordination space to assess the extent of local adaptation within each sampled population
- Quantify genomic offset per population
- Calculate neutral and adaptive genetic diversity per population based on standing neutral genetic diversity (nucleotide diversity) and local adaptation of individuals and populations
- Build adaptive SDMs using these locally adapted individuals to gain a more thorough understanding of differential responses amongst locally adapted populations
- Evaluate the landscape influence on populations (‘Landscape barriers’, using Circuitscape) to assess the potential for evolutionary rescue of isolated populations and spread of neutral/adaptive genetic diversity
- Combine multiple analyses to integrate Exposure, Adaptive sensitivity, Neutral sensitivity and Landscape barriers to make Population vulnerability predictions
- Create summary PDFs with completely transparent log files recording all steps within the toolbox